General Text Feature Extraction¶

Our mission¶
The mission is to automate job applications for everybody.
Pre-done work¶
I already did the following steps:
- Crawling all the links pointing on sites that show job offers
- Scraping each job offer with all information available
- Job name
- Company and link to its description site
- Job type (part-time/full-time/freelance etc.)
- Description (Stepstone uses html-parts to divide each section of the job offer)
- Saving all of the divided descriptions in a DataFrame (pandas)
- 1st column: headline
- 2nd column: text
- Cleaning the data
- Reducing the labels from about 5000 to 5 so that we have the following labels:
- company: a description of the company ("About us"-page)
- tasks: a description of the job offered (especially the tasks of the job)
- profile: a description of the skills needed to fulfill the job's requirements ("Your profile")
- offer: a description of what the company offers for the applicant ("What to expect", "What we offer to you")
- contact: contact details for the applicant (also including a hint whom to adress in the application)
- Reducing the labels from about 5000 to 5 so that we have the following labels:
- Preprocessing Pipeline
- In another notebook described in detail, I found a way to preprocess text data in different formats/hierarchies. Also, there are many different types of pipelines that can evolve by combining different techniques.
- NOW: Feature Extraction
- This task mainly consists of information extraction to provide enough information for the algorithms to seeparate our data for classification.
What this notebook covers¶
In this notebook, the goal is to extract as much information as we can get from text. In addition, this feature extraction pipeline is there to generalize the task for variable text data.
The data we use¶
We use a dataframe which I scraped from www.stepstone.de. The scraping process is in a separate jupyter notebook.
Pipeline¶
Making imports¶
At first, we need to import the libraries and tools (incl. model) we need to start.
import string
import json
import io
import copy
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
from sklearn.pipeline import FeatureUnion
from sklearn.linear_model import SGDClassifier
from sklearn.model_selection import GridSearchCV
from pprint import pprint
import time
import gensim
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.feature_extraction.text import CountVectorizer
from gensim.models.doc2vec import Doc2Vec, TaggedDocument
from gensim.models.wrappers import FastText
from gensim.test.utils import get_tmpfile
from gensim.models.callbacks import CallbackAny2Vec
from sklearn.base import BaseEstimator, TransformerMixin
import inspect
import os.path
from sklearn.decomposition import PCA
import sys
import Preprocessing as Preprocess
Helping functions¶
- Firstly, I am using the
makePrintandreleasePrintto set the print out function to a log file and unbind it again, respectively. - Secondly, the
sparse2listfunction converts a sparse matrix into a common Python list. We will see how that helps us. - Thirdly,
normalizeFeaturesprovides a way to scale numbers to a certain range we can specify. This can be chosen to normalize features we have. The formula looks the following: $z_i=\frac{x_i-\min(x)}{\max(x)-\min(x)}$
orig_stdout = sys.stdout
# Printing Progress into file
def makePrint(filename = "logs/log.log"):
f = open(filename, 'w+')
sys.stdout = f
def releasePrint():
sys.stdout = orig_stdout
def sparse2list(sparse_matrix, length):
sparse_array = sparse_matrix.toarray()
result = [[] for i in range(length)]
for i in range(length):
result[i] = list(sparse_array[i])
return result
def normalizeFeatures(features,specs):
try:
list_min, list_max = specs
features_min, features_max = (min(features),max(features))
return [((list_max-list_min)*(f-features_min)/(features_max-features_min))+list_min for f in features]
except Exception as e:
print(str(e))
print(features)
#return features
Word Embeddings¶
Methods¶
Word Embeddings are a great way to represent textual data. The features which are treated as a vector for each word or document (in this case: document) are calculated by a neural network. You can basically say that "it aims to quantify and categorize semantic similarities between linguistic items based on their distributional properties in large samples of language data." (https://en.wikipedia.org/wiki/Word_embedding)
In the end, you can either have a great visualization or a great way to do ome math with words:
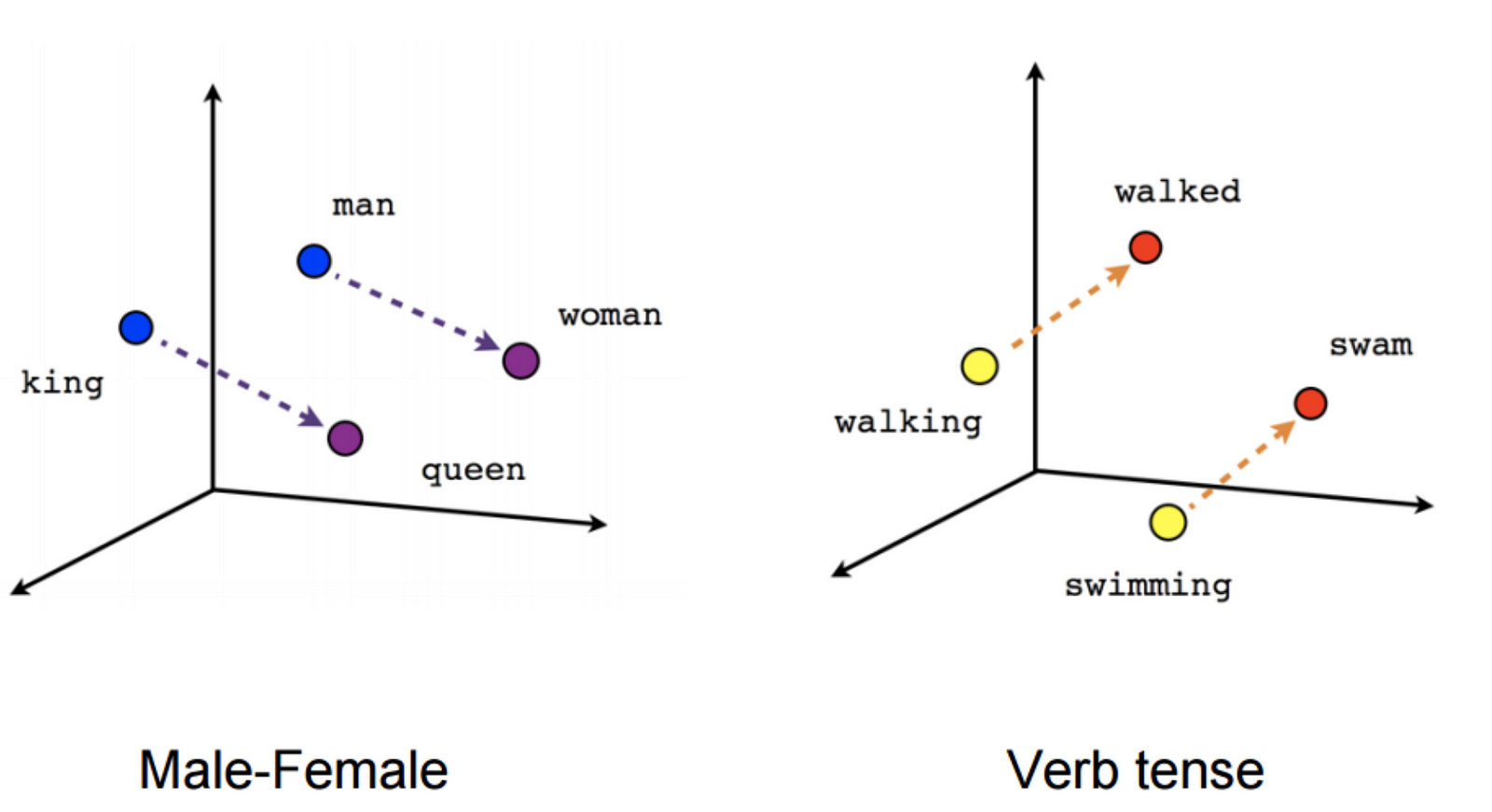
Source: https://cdn-images-1.medium.com/max/1600/1*jpnKO5X0Ii8PVdQYFO2z1Q.png
I use two different types of Embeddings:
Doc2Vec:
- This method has been developed on top of Word2Vec, which is one of the most common methods for Word Embeddings
- Research Paper: http://proceedings.mlr.press/v32/le14.pdf
FastText:
- I downloaded a freely available vector representation model from their website
- Unfortunately, we have to go our own way to featurize a whole document or abstract. I chose to get all the words' vector representation (300 features each) and used the mean of each document for missing words.
- FastText Website: https://fasttext.cc/
Classes¶
I then chose to develop classes that can be used by scikit learn's Pipeline feature to be able to build individual feature pipelines for different use cases. These classes are the following:
Doc2vec- This transformer takes in the documents that are supposed to be featurized by Doc2Vec and is building a model by given parameters. In order for the transformer to use a already built model, I integrated the parameter
finalto tell the class that the model doesn't have to be build but can be loaded from the disk. - In case the model has to be built,
newDoc2Vecis being called to do the task - Otherwise, if a model already exisits, the
Document2Vectorfunction can be used to turn the document into a feature vector
- This transformer takes in the documents that are supposed to be featurized by Doc2Vec and is building a model by given parameters. In order for the transformer to use a already built model, I integrated the parameter
getVectorRepresentation- This transformer is there to turn the word vectors based on the fastText model into a document vector.
- As I said before, non-existent words get a average-based feature representation (
Document2Vector) VectorExtractoris the underling function that can be constructed by a scikit learn Pipeline
class Doc2vec(BaseEstimator, TransformerMixin):
def __init__(self, vector_size = 50, window_size = 6,
min_count_for_words = 10, path_for_model = "doc2vec_", epochs = 40, final = False):
for arg, val in locals().items():
setattr(self, arg, val)
def fit(self, x, y=None):
return self
def transform(self, x, **transform_params):
if self.final:
self.path = self.path_for_model
else:
self.path = 'models/{path}_{vector_size}_{window_size}_{epochs}_{min_count_words}.wv'.format(path = self.path_for_model,
vector_size = self.vector_size,
window_size = self.window_size,
epochs = self.epochs,
min_count_words = self.min_count_for_words)
if not os.path.isfile(self.path):
print("Starting Doc2Vec Training: Model {}".format(self.path))
self.path = newDoc2Vec(x, vector_size=self.vector_size,
window_size=self.window_size,
min_count_for_words=self.min_count_for_words, path_for_model = self.path, epochs=self.epochs)
print("Using Doc2Vec Model: {}".format(self.path))
return [document2Vector(doc, embedding_type = "doc2vec", document_vectorizer = "mean", path_to_model = self.path) for doc in x]
def newDoc2Vec(X, vector_size = 50, window_size = 6,
min_count_for_words = 10, path_for_model = "doc2vec_", epochs = 40):
path = path_for_model
corpus = [TaggedDocument(doc, [i]) for i, doc in enumerate(X)]
model = Doc2Vec(
corpus,
size=vector_size,
window=window_size,
min_count=min_count_for_words,
workers=10)
model.train(corpus, total_examples = len(corpus), epochs=epochs)
model.save(path)
return path
class VectorExtractor(BaseEstimator, TransformerMixin):
def __init__(self, methods = ["Embedding"],
normalization = False,
embedding_type = "fastText",
pre_trained = True,
document_vectorizer = None,
lang = "de"):
for arg, val in locals().items():
setattr(self, arg, val)
self.methods = self.methods if self.methods else ["Embedding"]
# Further Options: embedding_type, document_vectorizer, tfidf_norm, use_idf, smooth_idf, sublinear_tf, lang
def fit(self, x, y=None):
return self
def transform(self, documents):
new_docs = copy.deepcopy(documents)
word_analyses = [[] for i in range(len(documents))]
if "Embedding" in self.methods or "WordFeatures" in self.methods:
for i in range(len(documents)):
if "Embedding" in self.methods:
for j in range(len(documents[i])):
doc = copy.copy(documents[i][j])
word_analyses[i].append([])
pre_trained = True if self.pre_trained != None else self.pre_trained
embedding_type = "fastText" if not self.embedding_type else self.embedding_type
document_vectorizer = "mean" if not self.document_vectorizer else self.document_vectorizer
lang = "de" if not self.lang else self.lang
word_analyses[i][j].extend(getVectorRepresentation(doc, pre_trained, lang, embedding_type))
if self.normalization:
new_docs[i] = normalizeFeatures(document2Vector(word_analyses[i], embedding_type), specs = (min(new_docs[i]), max(new_docs[i])))
else:
new_docs[i] = document2Vector(word_analyses[i], embedding_type)
if "WordFeatures" in self.methods:
new_docs[i].extend(normalizeFeatures(Preprocess.preprocessingOrchestrator("".join(documents[i]), include=["WordFeatures"]), specs = (min(new_docs[i]), max(new_docs[i]))))
for nr, doc in enumerate(new_docs):
if any([True if math.isnan(x) else False for x in doc]):
new_docs[nr] = [0.0]*len(new_docs[nr])
return new_docs
def getVectorRepresentation(token, pre_trained = True, lang = "de", embedding_type = "fastText"):
global fast_text_vector_model
global fast_text_average
global fast_text_vector_sum
global fast_text_vector_count
if embedding_type == "fastText":
if "fast_text_average" not in globals():
fast_text_vector_sum, fast_text_average, fast_text_vector_count = (0., 0., 0.)
vector = []
if not 'fast_text_vector_model' in globals():
print("Loading language model...")
fast_text_vector_model = FastText.load_fasttext_format('../{}_model.bin'.format(lang))
print("Language model loaded...")
else:
try:
vector = list(fast_text_vector_model[token])
fast_text_vector_sum += sum(vector)
fast_text_vector_count += 1
fast_text_average = fast_text_vector_sum / fast_text_vector_count
except:
vector = [fast_text_average for i in range(300)]
return vector
def document2Vector(word_vectors, embedding_type = "fastText", document_vectorizer = "mean", path_to_model = "doc2vec_"):
global doc2vec_vector_model
if embedding_type == "fastText":
if document_vectorizer == "mean":
return [sum(x)/len(word_vectors) for x in zip(*word_vectors)]
elif embedding_type == "doc2vec":
if not 'doc2vec_vector_model' in globals():
print("Loading language model...")
fname = path_to_model
doc2vec_vector_model = Doc2Vec.load(fname)
print("Language model loaded...")
doc = list(doc2vec_vector_model.infer_vector(word_vectors))
return [v.item() for v in doc]
Preprocessing Pipeline elements¶
These functions are existent to build a generalized feature pipeline that can be used by scikit learn.
Preprocessingis built upon the Preprocessing Pipeline I created in another notebook. You can choose different parameters to set up the transformer. This then uses thepreprocessingOrchestratorof the Preprocess module to perform the necessary steps. As I already described the different techniques before, I will not introduce the methods used by this module.Normalizeris the following part that is connected to the helper functionnormalizethat I described earlier. The class is used to provide the surroundings, such as data types and missing values.
class Preprocessing(BaseEstimator, TransformerMixin):
def __init__(self, include = [], langs = ["de"],
labels_separated = True,
unicode_seperator = True,
keep_emojis= False,
custom_abbreviations= [],
delete_abbreviations= [],
custom_stopwords= [],
stopword_level= 1,
n_gram_size= 2,
punctuation_level= 3,
punctuation_list = [],
which_special_char = []):
for arg, val in locals().items():
setattr(self, arg, val)
def fit(self, x, y=None):
return self
def transform(self, x, **transform_params):
res, y = Preprocess.preprocessingOrchestrator(x, y=[], **self.__dict__)
for nr, x in enumerate(res):
if len(x) == 0:
res[nr] = [""]
return res
def get_params(self, deep):
out = {
"include" : [],
"langs" : ["de"],
"labels_separated" : True,
"unicode_seperator" : True,
"keep_emojis" : False,
"custom_abbreviations" : [],
"delete_abbreviations" : [],
"custom_stopwords" : [],
"stopword_level" : 1,
"n_gram_size" : 2,
"punctuation_level" : 3,
"punctuation_list" : [],
"which_special_char" : []
}
for key in self._get_param_names():
value = getattr(self, key, None)
if deep and hasattr(value, 'get_params'):
deep_items = value.get_params().items()
out.update((key + '__' + k, val) for k, val in deep_items)
out[key] = value
return out
import math
class Normalizer(BaseEstimator, TransformerMixin):
def __init__(self, min_, max_, which, weight = 1):
self.min_ = min_
self.max_ = max_
self.which = which
self.weight = weight
def fit(self, x, y=None):
return self
def transform(self, documents):
new_docs = documents
if type(documents) != list:
new_docs = sparse2list(documents, documents.shape[0])
print("Transforming: ",self.which)
new_docs = [normalizeFeatures(document, specs = (self.min_, self.max_))*self.weight for document in new_docs]
for nr, doc in enumerate(new_docs):
if any([True if math.isnan(x) else False for x in doc]):
new_docs[nr] = [0.0]*len(new_docs[nr])
return new_docs
Defining helpers for metrics and logs¶
These functions help us to validate the preprocessing and model building success.
get_metrics¶
...is the function to measure the score for a model when it comes to classification. I took the following parameters for the evalutation step:
- The confusion matrix: One of the most common tools to see how much data has been classified to one group
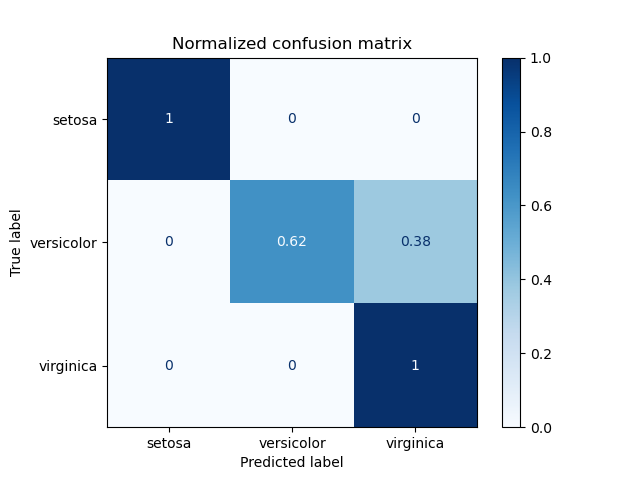 Source: https://scikit-learn.org/stable/_images/sphx_glr_plot_confusion_matrix_002.png
Source: https://scikit-learn.org/stable/_images/sphx_glr_plot_confusion_matrix_002.png
- Accuracy: This is the simplest one. It tells us how much percent has been classified correctly
- F1-Score: This is the mixture of the above metrics. It only works for binary data though (when you have trues and falses)
 Source: https://i.stack.imgur.com/U0hjG.png
Source: https://i.stack.imgur.com/U0hjG.png
where TP = True Positives (correctly classified Trues), TN = True Negatives (Falses classified as Trues), FP = False Positives (wrongly classified Trues), FN = False Negatives (correctly classified Falses)
Log class¶
- this class is supposed to keep track of all classifications that are done
- Storing this information helps us to gain information on what parameters have had a great impact on our classification results
def get_metrics(y_pred, y_true):
# compute more than just one metrics
chosen_metrics = {
'conf_mat': metrics.confusion_matrix,
'accuracy': metrics.accuracy_score,
'f1_score' : metrics.f1_score,
}
results = {}
for metric_name, metric_func in chosen_metrics.items():
try:
inter_res = metric_func(y_pred, y_true)
if type(inter_res) == np.ndarray:
results[metric_name] = inter_res.tolist()
else:
results[metric_name] = inter_res.item()
except Exception as ex:
inter_res = None
print("Couldn't evaluate {} because of {}".format(metric_name, str(ex)))
results['conf_mat'] = results['conf_mat']
return results
class Log():
def __init__(self, start):
self.start = time.time()
self.logs = {}
self.which = ""
def scorer(self, clf, X_val, y_true_val, feature_name = None, classifier = None):
if self.which not in self.logs:
self.logs[self.which] = []
# do all the work and return some of the metrics
y_pred_val = clf.predict(X_val)
results = get_metrics(y_pred_val, y_true_val)
if feature_name:
self.logs[self.which][feature_name][classifier] = []
self.logs[self.which][feature_name][classifier].append(results)
else:
self.logs[self.which].append(results)
return results['accuracy']
def save(self):
import pickle
path = "logs/LOG.log"
with open(path, 'wb') as handle:
pickle.dump(self, handle, protocol=pickle.HIGHEST_PROTOCOL)
Feature class¶
This class helps us to work with our newly designed features. The class has the following specifications:
preprocess_pipelineas attribute to be able to reconstruct the way the feature has been built. If it is successful, we can rebuild it the next time without trying different parameters,getPrePipelineget us the whole Pipeline- the
saveXfunction stores the features on the local disk to save RAM - the
getfunction retrieves the features from the disk and returns them as a list getTrainTestsplits the data in the common form of training and testing data. That way it is easier to deal with data splittingsaveResultsfunctions help us to keep the classification results inside the class in order to return them.getBestClassifiergives us the best classifier we can use to seperate the featured data
class Feature():
def __init__(self, name, X):
self.name = name
if type(X) == list:
self.dimension = len(X[0])
self.path = self.saveX(X)
else:
self.dimension = X.shape[1]
self.path = self.saveX(sparse2list(X, X.shape[0]))
self.clf = None
self.clf_name = None
self.clf_results = {}
self.clf_tuning_results = {}
self.preprocess_pipeline = None
self.transformer = None
def saveX(self, X):
print(self.name, type(X))
path = 'features/{}.json'.format(self.name)
with open(path, 'w') as outfile:
json.dump(X, outfile)
return path
def get(self):
temp = []
with open(self.path, 'r') as outfile:
temp = json.load(outfile)
return temp
def getTrainTest(self, percent):
x = self.get()
return x[0:round(percent*len(x))], x[round(percent*len(x)):]
def saveResults(self, clf, X_val, y_true_val, classifier = None):
y_pred_val = clf.predict(X_val)
results = get_metrics(y_pred_val, y_true_val)
return results
def saveResultsForParameterTuning(self, clf, X_val, y_true_val, classifier = None):
results = self.saveResults(clf, X_val, y_true_val, classifier)
# clf_tuning_results[0] = results, [1] = classifier itself
self.clf_tuning_results = (results, clf)
return results['accuracy']
def saveResultsForTesting(self, clf, X_val, y_true_val, classifier = None):
results = self.saveResults(clf, X_val, y_true_val, classifier)
if classifier not in self.clf_results:
self.clf_results[classifier] = []
# clf_results[clf][0] = results, [1] = classifier itself
self.clf_results[classifier].append((results, clf))
return results['accuracy']
def getBestClassifier(self, metric = 'accuracy'):
best = 0.0
best_estimator = None
for i, r in self.clf_results.items():
for e in r:
if e[0]["accuracy"] > best:
best_estimator = e[1]
best = e[0]["accuracy"]
if len(self.clf_tuning_results) and self.clf_tuning_results[0]["accuracy"] > best:
return self.clf_tuning_results[0]["accuracy"], self.clf_tuning_results[1]
else:
return best, best_estimator
def getPrePipeline(self):
return Pipeline([
('preprocess', self.preprocess_pipeline),
('transformer', self.transformer)
])
def __str__(self):
return str(self.__class__) + ": " + str(self.__dict__)
Corpus class¶
This class is the orchestrator of all the model finding and feature extraction work we can do. Here are the specifications:
- Storing data:
- The
Xdata: this is the raw text data (to save RAM, it gets deleted after we gathered the features) - The
ydata: the labels which we have to use to train classifier and evaluate them Doc2VecModel: the created model we can use for further task with our text datafeatures: The feature data in form of theFeatureclass
- The
- Preprocess and featurize data:
- Using predefined preprocessing pipelines, we can save the data as
preprocessedDoc - these preprocessedDocs get featurized by using tfidf, a very common way to vectorize the data
- Please note: WordEmbeddings like Doc2Vec use Tokenization as first step
- it is supposed to look like this:
[('preprocess', Preprocessing(custom_abbreviations=[], custom_stopwords=[], delete_abbreviations=[], include=['Tokenization', 'UnicodeCleaner', 'RemoveGermanChainWords', 'RemoveAbbreviations', 'ExpandCompoundToken', 'PunctuationFilter', 'ToLower', 'RemoveUmlauts', 'LanguageDetector'], keep_emojis=False, labels_separated=True, langs=['de'], n_gram_size=2, punctuation_level=3, punctuation_list=[], stopword_level=1, unicode_seperator=True, which_special_char=[])), ('transformer', Doc2vec(epochs=40, final=True, min_count_for_words=10, path_for_model='models/doc2vec.wv', vector_size=50, window_size=6))]
- Using predefined preprocessing pipelines, we can save the data as
- Evaluating the best classifiers:
- The
trainEntireModelfunction helps us to search for the best classifier for each Feature. It is using theParameterTestingmethod to do that (is described later on)
- The
- Train Combined Model:
- In order to combine the strenghts of different features, we can combine the features to get a more advanced model
- Be careful: Little RAM can be critical, it loads all features into the program
- In order to combine the strenghts of different features, we can combine the features to get a more advanced model
- Find the best Preprocessing Pipeline:
getBestPreprocessingPipelineuses the Feature Class and its functiongetBestClassifierto compare the results of the feature classification to finally return the best classifier based on accuracy.
- Transferring results to HDD:
saveAllis a special function to transfer the results, logs, feature data as well as models to a specific directory. Because I use the power of SSDs to process the data, I save it on my HDD to save space.
class Corpus():
def __init__(self, X, y, aimed_path, pre_paras):
self.X = X
self.y = y
self.path = aimed_path
self.preprocessedDocs = {}
self.pipelines = {}
self.Doc2VecModel = None
self.pre_paras = pre_paras
self.features = {}
self.level = 1
def preprocess(self, include, key):
## Preprocessing Step
self.preprocessedDocs[key], self.y = Preprocess.preprocessingOrchestrator(self.X, self.y, include, **self.pre_paras)
self.pipelines[key] = include
def makeFeatures(self, level = 2):
level = self.level
transformer = TfidfVectorizer(tokenizer = lambda x: x, preprocessor=lambda x: x)
doc2VecTransformer = Doc2vec(path_for_model = self.Doc2VecModel, final = True)
preTrainedEmbeddingTransformer = VectorExtractor(methods = ["Embedding"],
normalization = False,
embedding_type = "fastText",
lang = self.pre_paras["langs"][0])
for idx, pre_doc in self.preprocessedDocs.items():
self.features[idx] = Feature(idx,transformer.fit_transform(pre_doc, self.y))
self.features[idx].clf_name = testClassifier(self.features[idx], self.y, level = level)
self.pre_paras["include"] = self.pipelines[idx]
self.features[idx].preprocess_pipeline = Preprocessing(**self.pre_paras)
self.features[idx].transformer = transformer
print(self.features[idx].clf_name)
print(idx, "done")
self.features["Doc2Vec"] = Feature("Doc2Vec", doc2VecTransformer.fit_transform(self.preprocessedDocs["Tokenization"],
self.y))
self.features["Doc2Vec"].clf_name = testClassifier(self.features["Doc2Vec"], self.y, level = level)
self.features["PreTrainedEmbedding"] = Feature("PreTrainedEmbedding",
preTrainedEmbeddingTransformer.fit_transform(
self.preprocessedDocs["Tokenization"],
self.y))
self.features["PreTrainedEmbedding"].clf_name = testClassifier(self.features["PreTrainedEmbedding"],
self.y,
level = level)
self.pre_paras["include"] = self.pipelines["Tokenization"]
self.features["Doc2Vec"].preprocess_pipeline = Preprocessing(**self.pre_paras)
self.features["PreTrainedEmbedding"].preprocess_pipeline = Preprocessing(**self.pre_paras)
self.features["Doc2Vec"].transformer = doc2VecTransformer
self.features["PreTrainedEmbedding"].transformer = preTrainedEmbeddingTransformer
#del globals()
del CORPUS.preprocessedDocs
del X
print("Embeddings", "done")
print("Transformed to Features...")
def trainEntireModel(self, level):
level = self.level
for idx, features in self.features.items():
if features.clf_name:
features.clf = ParameterTesting(features, self.y)
def connectFeatures(self, level):
level = self.level
percent = 0.75
X_train, X_test = {}, {}
y_train, y_test = self.getLabels(percent)
clfs = {}
dimension = -1
for idx, features in self.features.items():
if features.clf is not None:
X_train[idx], X_test[idx] = features.getTrainTest(percent)
clfs[idx] = features.clf
if features.dimension < dimension or dimension == -1:
dimension = features.dimension
self.combinedPipeline = trainCombinedModel(X_train, y_train, X_test, y_test, clfs, dimension)
print("Max Dimensions: ", dimension)
def getLabels(self, percent):
y_train = self.y[:round(percent*len(self.y))]
y_test = self.y[round(percent*len(self.y)):]
return y_train, y_test
def getBestPreprocessingPipeline(self):
y_train, y_test = self.getLabels(0.75)
best = 0.0
best_feature = None
scorer = make_scorer(accuracy_score)
for idx, feature in self.features.items():
acc = feature.getBestClassifier()[0]
print(idx, "accuracy:", acc)
if acc > best:
best_feature = feature
best = acc
return best_feature.getPrePipeline()
def saveAll(self):
import os
newpath = r'D:/FeatureExtraction/{}'.format(self.path)
if not os.path.exists(newpath):
os.makedirs(newpath)
print(newpath+"/logs/")
os.makedirs(newpath+"/logs/")
os.makedirs(newpath+"/features/")
os.makedirs(newpath+"/models/")
copytree("C:/Users/Thorben/Documents/workspace/Applivo/Preprocessing/logs/",newpath+"/logs/")
copytree("C:/Users/Thorben/Documents/workspace/Applivo/Preprocessing/features/",newpath+"/features/")
copytree("C:/Users/Thorben/Documents/workspace/Applivo/Preprocessing/models/",newpath+"/models/")
Helper functions for saving data¶
def copytree(src, dst):
import shutil
import os
for item in os.listdir(src):
s = os.path.join(src, item)
d = os.path.join(dst, item)
if os.path.isdir(s):
shutil.copytree(s, d)
else:
shutil.copy2(s, d)
shutil.rmtree(src)
def makePaths():
import os
for p in ["C:/Users/Thorben/Documents/workspace/Applivo/Preprocessing/logs/",
"C:/Users/Thorben/Documents/workspace/Applivo/Preprocessing/features/",
"C:/Users/Thorben/Documents/workspace/Applivo/Preprocessing/models/"]:
if not os.path.exists(p):
os.makedirs(p)
Parameter Testing functions¶
While the train function is a method to quickly return accuracy results, the doGridSearch function makes use of the powerful parameter testing class GridSearchCV from scikit learn:
- You can vary parameters of each Transformer (even the ones we defined previously above)
- GridSearchCV uses k-fold cross-validation to evaluate the best parameters:
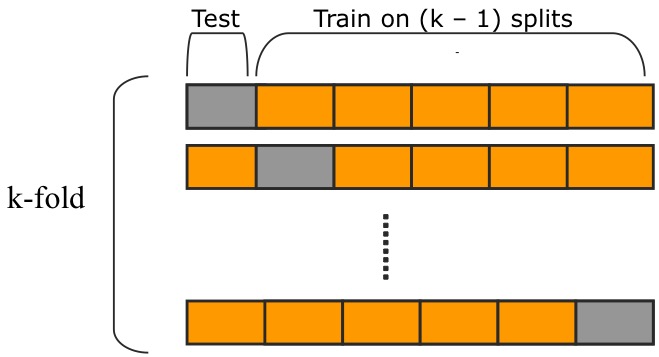 Source: https://raw.githubusercontent.com/qingkaikong/blog/master/2017_05_More_on_applying_ANN/figures/figure_1.jpg
Source: https://raw.githubusercontent.com/qingkaikong/blog/master/2017_05_More_on_applying_ANN/figures/figure_1.jpg- That way we can make use of the whole data that gets passed to the function
- the best parameters get stored and returned so we can assign them to our features
from sklearn import metrics
def train(classifier, X, y):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=33)
classifier.fit(X_train, y_train)
print("Accuracy: {}".format(classifier.score(X_test, y_test)))
return classifier
def doGridSearch(X, y, pipeline, parameters, k_folds = 2, printing = "grid_log.log", which = ""):
#makePrint(printing)
LOG.which = which
grid_search = GridSearchCV(pipeline, parameters, verbose=1, cv = k_folds, scoring = LOG.scorer)
print("Performing grid search...")
if type(pipeline) == "Pipeline":
print("Pipeline:", [name for name, _ in pipeline.steps])
print("Parameters:")
pprint(parameters)
t0 = time.time()
grid_search.fit(X, y)
print("Done in %0.3fs" % (time.time() - t0))
print()
print("Best score: %0.3f" % grid_search.best_score_)
print("Best parameters set:")
best_parameters = grid_search.best_estimator_.get_params()
for param_name in sorted(parameters.keys()):
print("\t%s: %r" % (param_name, best_parameters[param_name]))
#releasePrint()
print("Scoring the best model...")
LOG.logs[which].append(grid_search.cv_results_["params"])
print(LOG.logs)
results = {
"parameter": grid_search.cv_results_["params"],
"mean_train_score": grid_search.cv_results_["mean_train_score"],
"mean_test_score": grid_search.cv_results_["mean_test_score"]
}
return {
"best_pipeline": grid_search.best_estimator_,
"best_parameters": best_parameters,
"results": results
}
Doc2Vec Modelling¶
As seen above, we can construct a Word Embedding from our text data. This helps us to work with similar data in the future as well. The steps are the following:
- Searching for the best Doc2Vec parameters
- As we have seen in the GridSearch function, we can find out the best fitting parameters for the model by cross validating our model transformer and using different parameters for
vector_size,min_count_for_wordsandepochs - In this case, we are only using a small part of the text, so that the computational cost isn't that high
- As we have seen in the GridSearch function, we can find out the best fitting parameters for the model by cross validating our model transformer and using different parameters for
- Receiving the best parameters for the model
- After using a standardized classifier we use the accuracy metric to find out the best parameters
- Train a whole model
- The next step is to use all the text data to create a Word Embedding that we can use for further investigation and calculations
def getBestDoc2VecModel(X, y, level = 2):
length = min(len(X)/(4-level),1000)
X_testing = X[:length]
y_testing = y[:length]
parameter = {
# 12 tries
1 : {
"doc2vec__vector_size" : (50,100),
"doc2vec__min_count_for_words" : (5,10),
"doc2vec__epochs" : (100,150,200)
},
# 36 tries
2 : {
"doc2vec__vector_size" : (50,100,150),
"doc2vec__min_count_for_words" : (5,7,10),
"doc2vec__epochs" : (100,150,200,250)
},
# 60 tries
3 : {
"doc2vec__vector_size" : (50,100,150,200),
"doc2vec__min_count_for_words" : (5,7,10),
"doc2vec__epochs" : (100,150,200,250,300)
},
# 180 tries
4 : {
"doc2vec__vector_size" : (50,100,150,200,250),
"doc2vec__min_count_for_words" : range(5,11),
"doc2vec__epochs" : (100,125,150,175,200,250)
}
}
#### Building own Word Embedding
embedding_pipeline = Pipeline([
('doc2vec', Doc2vec(path_for_model = "doc2vec")),
('clf', SGDClassifier(random_state = 33))
])
grid_search_results = doGridSearch(X_testing, y_testing, embedding_pipeline, parameter[level],
printing = "logs/Doc2Vec_Evaluation.log",
which = "Doc2Vec_Evaluation")
epochs = grid_search_results["best_parameters"]["doc2vec__epochs"]
min_count_for_words = grid_search_results["best_parameters"]["doc2vec__min_count_for_words"]
vector_size = grid_search_results["best_parameters"]["doc2vec__vector_size"]
#model_building_log["Doc2Vec_Evaluation"] = grid_search_results["results"]
LOG.logs["Doc2Vec_Best_Model"] = {
"vector_size": vector_size,
"min_count_for_words": min_count_for_words,
"epochs": epochs,
#"accuracy": ,
}
print("Training best Word2Vec model with best parameters...")
best_model = newDoc2Vec(X, epochs = epochs,
min_count_for_words = min_count_for_words,
vector_size = vector_size,
path_for_model = "models/doc2vec.wv")
return best_model
Searching for the best classification algorithm¶
In order to create a model in the end, we need to make sure that we are using a classifier that does the work we expect. In consequence, we try out different sets of classifier depending on the level of detail (we come to that later). As before, we are using the accuracy metric to find out which classifier performs best for each feature that we feed the function with.
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.linear_model import Perceptron
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.metrics import accuracy_score
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import f1_score
from sklearn.metrics import make_scorer
def testClassifier(feature, y, level = 2):
X = feature.get()
keep_data = (level*0.2)+0.15
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=1-keep_data, random_state=33)
parameter = {
1 : {
"Stochastic Gradient Descent": SGDClassifier(),
"Logistic Regression": LogisticRegression(),
"Naive Bayes": MultinomialNB(),
},
2 : {
"Stochastic Gradient Descent": SGDClassifier(),
"Logistic Regression": LogisticRegression(),
"Naive Bayes": MultinomialNB(),
"Gradient Boosting": GradientBoostingClassifier(),
},
3 : {
"Stochastic Gradient Descent": SGDClassifier(),
"Logistic Regression": LogisticRegression(),
"Naive Bayes": MultinomialNB(),
"Gradient Boosting": GradientBoostingClassifier(),
"Perceptron": Perceptron(),
},
4 : {
"Stochastic Gradient Descent": SGDClassifier(),
"Logistic Regression": LogisticRegression(),
"Naive Bayes": MultinomialNB(),
"Gradient Boosting": GradientBoostingClassifier(),
"Perceptron": Perceptron(),
}
}
scorer = feature.saveResultsForTesting
best = 0.0
best_classifier = None
for idx, p in parameter[level].items():
clf = p
try:
clf.fit(X_train, y_train)
acc = scorer(clf, X_test, y_test, classifier = idx)
print(idx, "accuracy:", acc)
if acc > best:
best_classifier = idx
best = acc
except Exception as e:
print(str(e), " => occurred during choice of best classifier for", idx)
return best_classifier
Detailed classification evaluation¶
Now that we have found the best classifier, we want to adjust the classifier to get the best result. As we've done before, we use doGridSearch for trying out parameters that again depend on the level of detail defined for the Corpus class
As we get the best results based on accuracy, the tuning is over and we assign the classifier object to the feature we did the tuning for.
def ParameterTesting(feature, y, level = 2):
classifier = feature.clf_name
X = feature.get()
feature_name = feature.name
keep_data = (level*0.2)+0.15
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=1-keep_data, random_state=33)
clfs = {
"Stochastic Gradient Descent" : {
"classifier": SGDClassifier(),
"parameter": {
'loss': ('hinge', 'log', 'modified_huber', 'squared_hinge', 'perceptron',
'squared_loss', 'huber', 'epsilon_insensitive', 'squared_epsilon_insensitive'),
'alpha': (0.0001, 0.00001, 0.000001),
'penalty': (None, 'l2', 'l1', 'elasticnet')
}
},
"Naive Bayes" : {
"classifier": MultinomialNB(),
"parameter": {
'var_smoothing': (1e-6, 1e-7, 1e-8, 1e-9, 1e-10, 1e-11)
}
},
"Logistic Regression" : {
"classifier": LogisticRegression(),
"parameter": {
'penalty': ('l2', 'l1'),
'C': tuple(list(np.arange(0.8,1.2,0.1)))
}
},
"Gradient Boosting": {
"classifier": GradientBoostingClassifier(),
"parameter": {
'n_estimators': tuple(list(np.arange(80,200,20))),
'max_depth': tuple(list(np.arange(2,5,1)))
}
},
"Perceptron" : {
"classifier": Perceptron(),
"parameter": {
'penalty': (None, 'l2', 'l1', 'elasticnet'),
'alpha': (0.0001, 0.00001, 0.000001)
}
}
}
scorer = feature.saveResultsForParameterTuning
LOG.which = "Classifier_Parameter_Tuning"
grid_search_results = doGridSearch(X_train, y_train, clfs[classifier]["classifier"], clfs[classifier]["parameter"],
printing = "logs/Classifier_Parameter_Tuning.log",
which = feature_name)
clf = grid_search_results["best_pipeline"]
try:
acc = scorer(clf, X_test, y_test, classifier = "GridSearch For Best Accuracy")
print(clf, "accuracy:", acc)
except Exception as e:
print(str(e), " => occurred during choice of best classifier for", feature_name, "on", clf)
return grid_search_results["best_pipeline"]
Train combined model¶
The Corpus class leaves space for a combined model. The combined model has the purpose to represent each feature contained by the Corpus and building a model by combining these in a whole. The following steps are taken:
- We merge the data into one object
- We build an own pipeline for each of the features and also use
ItemSelectorto be able to distinguish the data between the features - We set different weights for each feature and try them out for the pipeline
- Finally, we return the model with the weights that led us to the best accuracy
Watch out: Because we load in the features all at once, we have to be aware that the RAM might not be able to handle it. This produces a MemoryError
class ItemSelector(BaseEstimator, TransformerMixin):
#key corresponding to the desired value in a mappable.
def __init__(self, key):
self.key = key
def fit(self, x, y=None):
return self
def transform(self, data_dict):
return data_dict[self.key]
def trainCombinedModel(X_train, y_train, X_test, y_test, clf, dim):
pipeline = []
starting_weights = {}
parameter_weights = []
parameter = {"feature_union__transformer_weights":[]}
for idx, x in X_train.items():
pipeline.append(('{}_Pipeline'.format(idx), Pipeline([
('selector', ItemSelector(key=idx)),
('PCA', PCA(n_components = min(len(x[0]),dim)))
])))
starting_weights['{}_Pipeline'.format(idx)] = 1
parameter_weights = [starting_weights]
for idx, w in starting_weights.items():
temp = copy.copy(starting_weights)
temp['{}_Pipeline'.format(idx)] = 0
parameter_weights.append(temp)
parameter["feature_union__transformer_weights"] = parameter_weights
scorer = LOG.scorer
LOG.which = "Combined_Feature_Model"
best_acc = 0
best_pipeline = None
if "Combined_Feature_Model" not in LOG.logs:
LOG.logs["Combined_Feature_Model"] = {}
for i, pm in enumerate(parameter_weights):
LOG.logs["Combined_Feature_Model"]["Combined_{}".format(i)] = {}
end_pipeline = Pipeline([
('feature_union', FeatureUnion(
pipeline, transformer_weights = pm
)),
('clf', SGDClassifier(random_state = 33))
])
end_pipeline.fit(X_train, y_train)
acc = scorer(end_pipeline, X_test, y_test, feature_name = "Combined_{}".format(i), classifier = "SGD")
if acc > best_acc:
best_pipeline = end_pipeline
best_acc = acc
print("Combined => ", i, "accuracy:", acc)
return best_pipeline
Preprocessing Pipeline Generator¶
This function exists to hand over the preprocessing steps determined by the level of detail. The higher the level, the higher the variety of preprocessing steps. This is a good tool to have different features on the one hand, and to be able to rank different preprocessing pipelines on the other hand.
def getPreprocessingSteps(level, emojis, force_language):
preprocessingSteps = OrderedDict()
general_steps = ['UnicodeCleaner', 'RemoveGermanChainWords', 'RemoveAbbreviations',
'ExpandCompoundToken', 'PunctuationFilter', 'ToLower', 'RemoveUmlauts']
tokenization_iteration_levels = [[], ["Tokenization", "POSTagging"],
["Tokenization", "POSTagging", "Stopwords"],
["Tokenization", "POSTagging", "Stopwords", "StemText", "FineGrainedPOSTagging"]]
# Tokenization
preprocessingSteps["Tokenization"] = ['Tokenization']
# lemmatization
preprocessingSteps["Lemmatization"] = ['Lemmatization']
# Stemming
preprocessingSteps["StemText"] = ['Tokenization','StemText']
# POSTagging
preprocessingSteps["POSTagging"] = ['POSTagging']
# POSTagging/FineGrained
preprocessingSteps["FineGrainedPOSTagging"] = ['FineGrainedPOSTagging']
# POSTagging/FineGrained
preprocessingSteps["Stopwords"] = ['Tokenization','Stopwords']
for key, step in preprocessingSteps.items():
if key in tokenization_iteration_levels[level]:
step.extend(general_steps)
if force_language:
step.append("LanguageDetector")
if emojis:
step.append("HandleEmojis")
yield key, step
Core¶
The ModelBuilding function actually combines all the methods that are discussed above. You only have to set a few parameters:
name: The name determines in which directory the results are getting stored after the whole procedure. It is important to pick another name for each task so that the files don't get overwritten.task: This is the parameter that tells the program which results to return. You can choose between 3 different scenarios:- PreprocessingPipeline (default): this receives the features you want and also returns the connected classifier that performs best. That way you can already determine the best feature for classification, though you haven't found the best parameters yet
- GetEmbedding: this trains the Word Embedding model with the best parameters and returns a string with the path to the saved model
- GetBestModels: This function does the same thing as PreprocessingPipeline but also determines the best parameters for each classifier as well as the combined model. I actually deactivated the last part because I got the memory error last time.
force_language: This is the parameter to sort out every text in a language that is not specified in thelanguagesparameter. Of course, this can sometimes lead to mistakenly erase a text, but the accuracy is at 98%.emojis: If there are emojis in the text, this parameter helps to handle them in the preprocessing steps.detail_levels: This is the parameter I have been talking about earlier. It helps to set a limit for computational expenditure
from collections import OrderedDict
def ModelBuilding(X, y_, name, task = "PreprocessingPipeline", force_language = True, languages = ["de"], emojis = False,
detail_levels = {
"tokenization": 1,
"embeddings": 1,
"corpus": 1
}):
global LOG
global CORPUS
if "LOG" in globals() and "CORPUS" in globals():
del globals()["LOG"]
del globals()["CORPUS"]
LOG = Log(time.time())
start = time.time()
makePaths()
pre_paras = {
"langs" : languages,
"labels_separated" : True,
"unicode_seperator" : True,
"keep_emojis" : False,
"custom_abbreviations" : [],
"delete_abbreviations" : [],
"custom_stopwords" : [],
"stopword_level" : 1,
"n_gram_size" : 2,
"punctuation_level" : 3,
"punctuation_list" : [],
"which_special_char" : []
}
CORPUS.level = detail_levels["corpus"]
CORPUS = Corpus(X, [[n_y] for n_y in y_], aimed_path = name, pre_paras = pre_paras)
for key, step in getPreprocessingSteps(level = detail_levels["tokenization"],
emojis = emojis, force_language = force_language):
## Preprocessing Step
print("step", step)
CORPUS.preprocess(key = key, include = step)
## Building a word embedding for the documents
if key == "Tokenization":
CORPUS.Doc2VecModel = getBestDoc2VecModel(CORPUS.preprocessedDocs[key], CORPUS.y,
level = detail_levels["embeddings"])
if task == "GetEmbedding":
break
CORPUS.makeFeatures(level = 4)
if task == "PreprocessingPipeline" or task == "GetEmbeddding":
res = CORPUS
LOG.save()
CORPUS.saveAll()
return res
elif task == "GetBestModels":
CORPUS.trainEntireModel(level = 1)
#CORPUS.connectFeatures(level = 1)
res = CORPUS
LOG.save()
CORPUS.saveAll()
return res
print("Time elapsed:", time.time() - start, "sek.")